DNA

What is DNA?
Advancements in our understanding of the basic building blocks of life has led to an exponential increase in the widespread awareness of DNA… but how well do you really know DNA?
You probably know that differences in DNA directly determine the differences between you and I, between you and your pet or the microbes living inside you, and even between your plants you lovingly called Clara and Phyll.
DNA is a molecular code or template that when “read,” instructs cells of the body on what to make, how to make it, and how to behave. Information in the DNA code contains Genes, which perform unique functions in your cells.
Genes and Variants
Within a species we all have the same Genes but the exact code in them can be different. Differences of the same Genes are called alleles. Some you can see, such as eye color and hair color.
Sometimes alleles known as variants can lead to problems in your body that you can’t see. Those problems may affect your health, response to foods, and response to drugs.


What is important about your Genes?
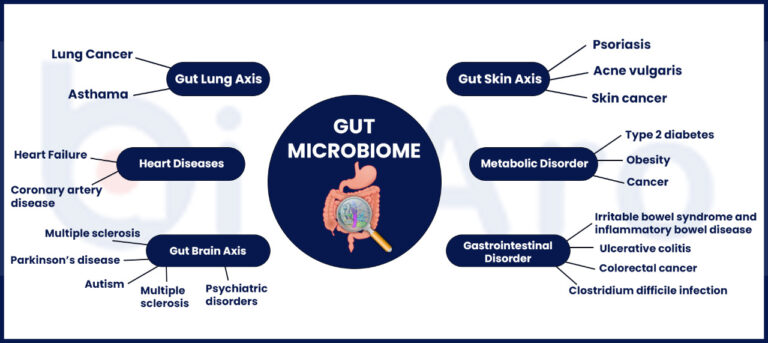
Genetic testing looks for alleles or variants that may change the activity of your immune system and how your body uses food and drugs. These variants may impact lifestyle and change disease risk.
Genetic testing is useful in many areas of medicine and can change the medical care you or your family member receives to get you the best treatment possible.
Genome and Exome

What is the Genome and Exome?
When someone references a genome they are referring to the entire DNA code of a person, which includes Genes and other DNA. All of your chromosomes from both parents are considered in this and it includes 3 billion nucleotides. If nucleotides were letters being read, it would be over 1000 copies of War and Peace in length.
While we have a lot of DNA code, not all of it encodes our Genes. Only ~1-2 % of our genome is “coded” to make the Genes that we are familiar with and these coded sequences are called the exome.
The exome captures much of the variants that we associate with diseases, which is why many scientists analyze the Whole Exome Sequence (WES) and not the Whole Genome Sequence (WGS). However, since improvements in technology have enabled more whole genomes to be sequenced, we are seeing diseases that associate with regions in the genome that are “intergenic” or between Genes and coding areas. Chromosome structures and intergenic regions have important roles to play in the function of a healthy cell.
Therefore, the genome comprises everything, the exome is a subset of “everything” and both are becoming increasingly valuable in health research and diagnostics.
Next Generation Sequencing

The DNA code is represented by the first letter of the base of the nucleotide molecule in the sequence: A for adenine, T for thymine, C for cytosine, and G for guanine. A stretch of DNA is called an oligonucleotide and would be represented by these bases to make a “word”. You can think of our chromosomes as very, very long words.
As was mentioned earlier, we have differences in our codes to cause different alleles and variants in our bodies. Consider it to be a string of ATCGs that are “spelled” differently with letter changes that change the letters in the word for different ones (A to T), delete letters, add letters, and copy and paste groups of letters to different parts of chromosome or onto another. The changes arose from mutations in the histories of our families and were propagated by our ancestors until we were born. We are at least 50 % likely to have that change in our children too (if you and your partner are from the same population, there is a greater chance you share ancestral Genes).
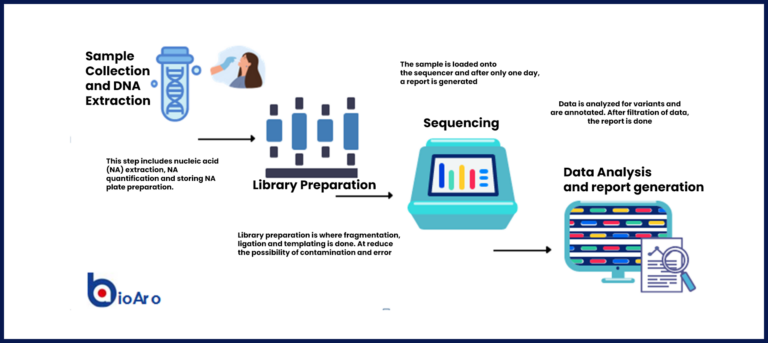
TDNA codes are “read” by the laboratory technique known as sequencing that allows the scientists to identify each base in a sequence. The field of study started by being able to identify only short reads of one type of DNA sequence at a time (think of lots of copies of the same word ). It then progressed to be able to identify many words at a time. This is what is known as Massive Parallel Sequencing (MPS) or the more commonly known term: Next Generation Sequencing (NGS).
NGS technology started with only selecting targeted parts of the genome to look at for determining if there is a genetic cause or contribution to a certain disease. Now, the technology has grown to allow sequencing of the whole exome and whole genome.